Medical literature has been rapidly expanding throughout the last few decades, where the number of yearly publications has an exponential growth since early 2000.1 Processing and understanding what is being published and analyzing the topics that are considered “hot topics” is now more difficult to be done manually. However, there has been a parallel development in automatic detection techniques and natural language processing (NLP). NLP is a machine learning technique involving a set of methods and computer-aided algorithms designed to detect patterns in textual data.2 With the exponential growth of medical data, NLP has the potential to establish itself as fundamental in every aspect of the healthcare industry due to the necessity of more efficient and accurate methods of analyzing and utilizing this information. NLP algorithms can process and understand vast amounts of unstructured medical text data, such as electronic health records, medical notes, and research papers.3 It has the potential to be used in automating routine tasks, which reduces errors and streamlines the workflow, improving patient outcomes, lowering costs, and increasing the efficiency of the healthcare industry. Topic detection, in the context of NLP, can be defined as an algorithm that automatically identifies topics based on the content of a scientific article.4 Topic detection algorithms analyze large amounts of text data and identify the underlying themes or topics present in the data. Topic detection can jumpstart the process of creating specific, topic-based databases by eliminating the time-consuming manual labor that has previously stifled its progress,4 enabling physicians to remain informed about the latest developments in their respective fields. In addition, topic detection can also be used to identify trends in literature, providing valuable insight into the direction of the field, which may be useful to editorial boards in assembling a qualified pool of editorial members or reviewers by identifying topics that are receiving the most attention and investment in the field or guiding researchers to areas with gaps in literature where their efforts may be impactful. The purpose of this research article was to develop an NLP-based method for extracting topics from abstracts. Using such a novel model, we assessed the main topics of published articles by top medical journals in the last three years. Such an assessment will provide valuable insight to researchers and funders about current research trends and guide future work in these fields. We will also provide the details of this novel and robust methodology that can be used by other researchers from different fields and sub-fields of medicine.
The current study composed of two main parts, a bibliometric method to extract original articles published by top journals, followed by a NLP method to classify topics discussed by extracted articles.
We chose the top non-specialized medical journals in the 2021 Journal Citation Report®, with an impact factor (IF) above 50. These journals included Lancet (IF 202.731), New England Journal of Medicine (IF 176.082), Journal of the American Medical Association (IF 157.375), and British Medical Journal (IF 96.216). We did not include journals that publish reviews (i.e., Nature Reviews Disease Primers). All included journals were categorized under the “Medicine, General & Internal – SCIE” category.
We used Web of Science to extract the data on the 7th of January 2023, where we searched for all articles published in the aforementioned journals in the years 2020, 2021, or 2022. We restricted the search for original articles. We excluded non-original articles that were included even after applying the search query restriction (e.g., clinical cases published in NEJM). For each article, we included details about its journal, year, authors, affiliations, and funding body. In addition, we obtained the number of citations received according to the Web of Science database up to the date of search. We also included each article’s abstract, which will be further used as input to the NLP model to classify the articles. We manually excluded article types that did not have an abstract and did not report original data, including JAMA performance improvement, analysis in BMJ, updates on systematic review in BMJ, and special reports in NEJM without original data.
Transformers are a class of deep learning architectures that were introduced by Vaswani et al in 2017. They use self-attention mechanisms to attend to different parts of the input during training and inference which makes it particularly strong in NLP. BERT (Bidirectional Encoder Representations from Transformers) is one of the well-known transformer models for its benchmark performance in a variety of NLP tasks.5
To find the most prevalent themes in our dataset, we utilized BERTopic, a topic modeling approach based on the BERT language model. Modern topic modeling techniques like BERTopic use language models that have already been trained to capture the semantic meaning of words and sentences. Topic representations in BERTopic are generated in three steps, each document is first transformed using a trained language model into its embedding representation, second, the dimensionality of the generated embeddings is then decreased prior to clustering to improve the clustering procedure, and finally, the topic representations are retrieved from the document clusters using a customized class-based form of TF-IDF (term frequency-inverse document frequency).5
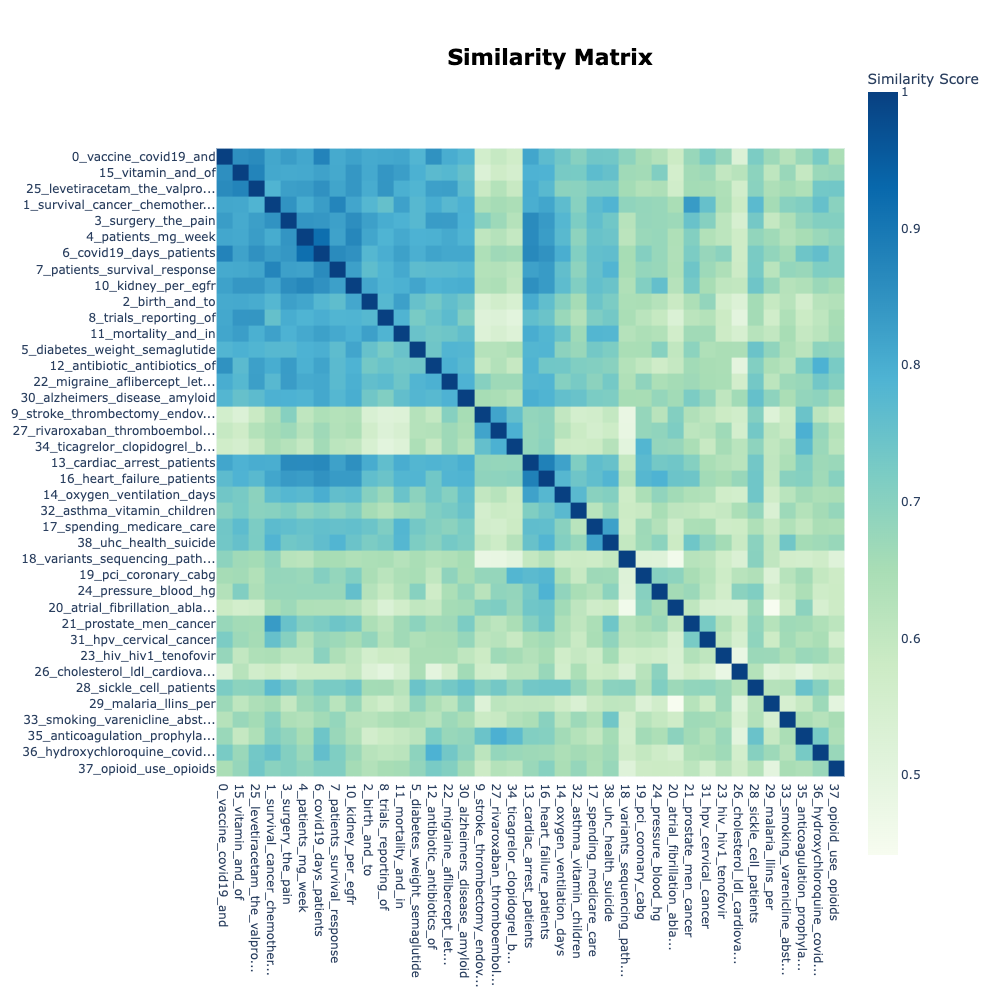
To cluster the documents in our dataset into different topics we used the default settings for the open-source BERTopic model, which includes using the Cosine Similarity measure to calculate the similarity between documents. The values for cosine similarity range between -1 and 1, with 1 denoting that the documents are identical, 0 denotes that they are orthogonal, and -1 for being diametrically opposed, the values correspond to the angle between the vectors. Consequently, BERTopic generates a similarity matrix to express the degree of similarity between every pair of texts in the dataset6 (figure 2).

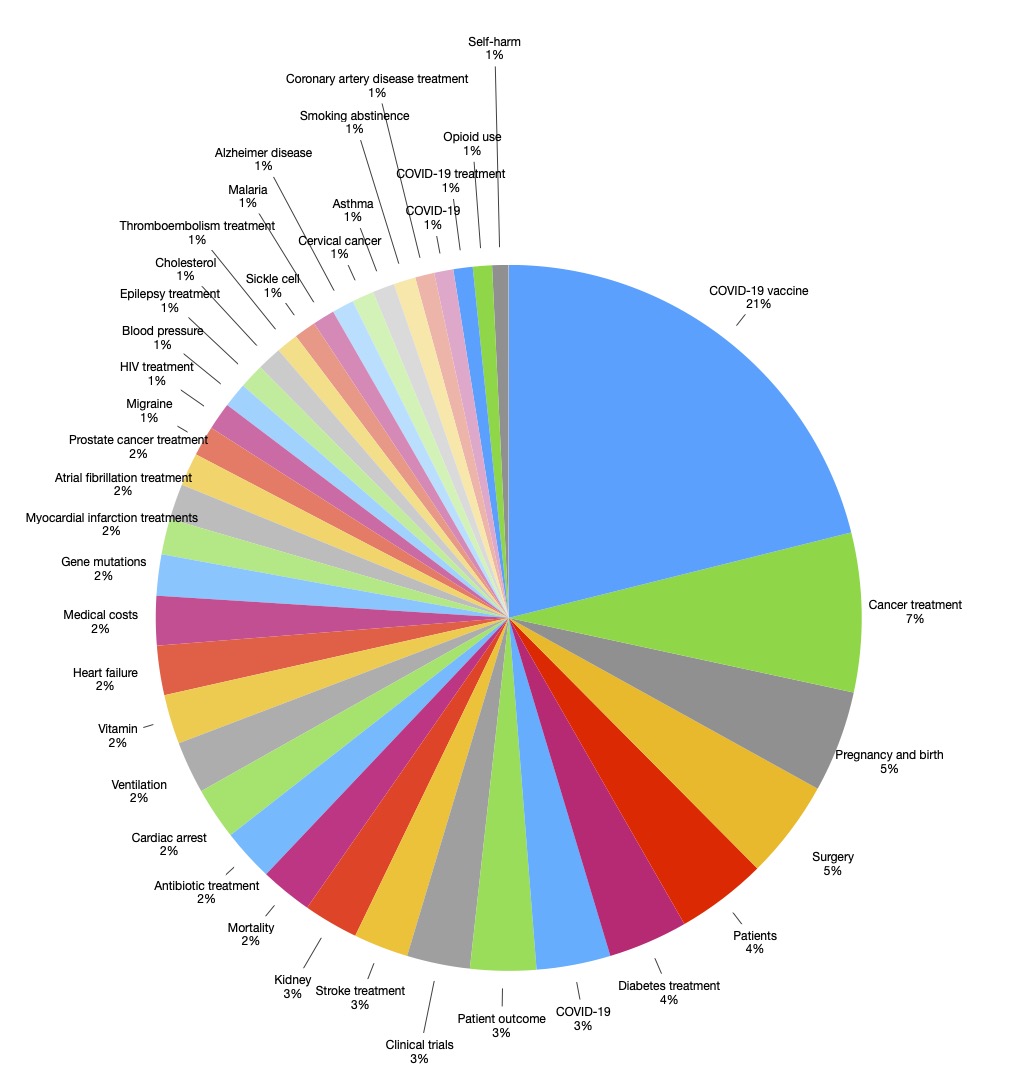
The model provided 39 different topics, with each having five keywords to describe it. Three physicians were consulted to provide a descriptive title for each topic, and the consensus description from the three physicians was adopted. Discrepancy between physicians mandated collaborative discussions to end up with an agreed upon description. After that, we categorized topics into main specialties.
We used IBM SPSS Statistics for Windows, version 26.0 (IBM Corp., Armonk, N.Y., USA). in our analysis. We used mean (± standard deviation) to describe continuous variables. We used count (frequency) to describe other nominal variables. We compared the number of articles published by each journal on each topic using chi-square test. One-way ANOVA for different bibliometric metrics (i.e., citations, references, authors, and pages) between the four journals, and we reported the results using mean and standard deviation. We adopted a p-value of 0.05 as a significant threshold.
The initial list of articles from included journals comprised a total of 2,081 articles. 144 articles were excluded according to the described criteria, leaving a total of 1,937 included articles after manual exclusion. The majority were for NEJM (705, 36.4%), followed by Lancet (448, 23.1%), JAMA (419, 21.6%), and BMJ (367, 18.9%).
The model was able to categorize 1,540 of the publications (79.42%) into 39 different topics. The most common topics published in top medical journals were involving the COVID-19 vaccine, cancer treatments, pregnancy, and birth. Figure 1 shows the 39 topics published by top medical journals and the proportion of publications in each topic.
Despite separating the topics into 39 distinct ones, some of these topics were related and were discussed in the context of each other. The model generated a similarity matrix that shows the degree of similarity between topics. A high similarity index (i.e., above 80%) was found between COVID-19 vaccine and vitamin (85.5%); prostate cancer and survival (83.4%); COVID-19 vaccine and antibiotic (85%); COVID-19 vaccine and COVID-19 (87.7%); COVID-19 and cardiac arrest (86.6%); surgery and cardiac arrest (86.2%); cancer treatment and kidney (82%); COVID-19 and mortality (82.4%).
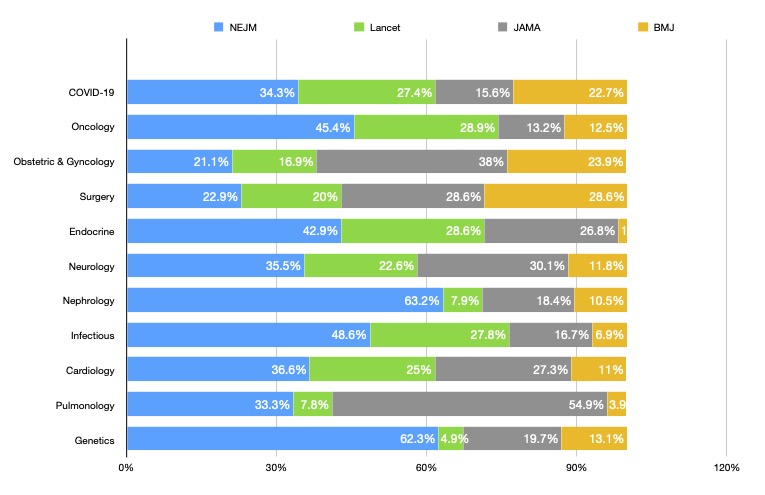
According to field categorization, 11 different fields resulted. Figure 3 shows the proportion of articles contributed by each journal in each field. The difference in topic distribution was significantly different between journals (p< 0.001).
A significant difference between top journals in respect to:
The number of citations received (p< 0.001), with Lancet receiving the highest mean (265.36 ±1224.04) and BMJ receiving the lowest mean (64.34 ±202.31) citations.
The number of references in each article (p<0.001), with BMJ articles having the highest number of references (42.6 ±23.45) and NEJM having the lowest mean (29.39 ±9.84) number of references.
The number of authors in each article (p< 0.001), with Lancet having the highest mean number of authors (61.03 ±212.93) and BMJ having the lowest mean number of authors (14.38 ±11.21).
The number of pages (p< 0.001), with Lancet having the highest mean number of pages (11.96 ±4.65) and JAMA having the lowest mean number of pages (10.39 ±1.77).
Table 1 details the mean number of citations, references, authors, and pages for articles published in top medical journals.
| N | Mean | Std. Deviation | |||
|---|---|---|---|---|---|
| References in each article | NEJM | 705 | 29.39 | 9.842 | <0.001 |
| Lancet | 448 | 37.96 | 23.303 | ||
| JAMA | 419 | 30.86 | 8.826 | ||
| BMJ | 367 | 42.60 | 23.450 | ||
| Total | 1939 | 34.19 | 17.566 | ||
| Citation according to Web of Science | NEJM | 705 | 243.93 | 935.012 | <0.001 |
| Lancet | 448 | 265.36 | 1224.041 | ||
| JAMA | 419 | 94.26 | 349.420 | ||
| BMJ | 367 | 64.34 | 202.307 | ||
| Total | 1939 | 182.55 | 839.350 | ||
| Citations in all databases | NEJM | 705 | 252.08 | 999.733 | <0.001 |
| Lancet | 448 | 279.07 | 1314.041 | ||
| JAMA | 419 | 96.59 | 355.416 | ||
| BMJ | 367 | 66.19 | 209.300 | ||
| Total | 1939 | 189.53 | 897.148 | ||
| Number of pages | NEJM | 705 | 10.93 | 1.960 | <0.001 |
| Lancet | 448 | 11.96 | 4.651 | ||
| JAMA | 419 | 10.39 | 1.769 | ||
| BMJ | 367 | 11.08 | 3.440 | ||
| Total | 1939 | 11.08 | 3.096 | ||
| Number of authors | NEJM | 705 | 24.56 | 20.775 | <0.001 |
| Lancet | 448 | 61.03 | 212.927 | ||
| JAMA | 419 | 20.60 | 18.984 | ||
| BMJ | 367 | 14.38 | 11.207 | ||
| Total | 1939 | 30.20 | 104.948 |
We developed a novel method based on the newly developed NLP model BERTopic.5 This method can be used to analyze the topics discussed by a large number of articles, where the model can have articles’ abstract as the input, and it will provide a number of valuable outputs, including the most common topics discussed, their relation to each other, and the similarity between them. We applied this novel method to articles published by general medical journals in the last three years. We found that almost 80% of published articles were related to 39 distinct topics in 11 fields. We also found that each journal focused on certain topics more than others, rather than being general. For example, JAMA published the highest percentage of pulmonology-related articles compared to other journals, while NEJM published the highest number of nephrology and genetics-related articles. We also performed a bibliometric analysis for the included articles, showing significant differences between journals regarding the number of citations, and number of authors per article, along with several references and pages in each article. We believe such methodology can be applied to other specialties and journals, helping researchers and policymakers understand the fields that are currently well-published and the topics that are now being researched.
Approximately 25% of all the papers published in these 4 journals during 2020-2022 were related to COVID-19 and its treatment. With the preceding global COVID-19 pandemic, this was not surprising. This rapidly increasing momentum that COVID-19-related articles had in literature has been described as one of the biggest explosions of scientific literature ever, with >450000 articles published within a year of the emergence of the disease.7 Despite the large percentage of papers being sorted into COVID-19, some topics, such as Vitamins, showed a very high degree of similarity with COVID-19, which could mean that the proportion of studies regarding COVID-19 and its treatment is underestimated as these studies could be describing the same topic. The topics published continue to follow an expected trend, with more common entities having more papers published on them. Cancer is the second most common cause of death worldwide (WHO), and 7% of all papers published in these journals have been about the treatment of different cancers. Our model was able to identify these topics among 1500 papers, this can potentially be replicated on a larger scale with more papers, more journals, and over a longer period of time. The ability of the model to do so with minimal manual labor means that identifying trends in literature becomes a much easier and more efficient task. Trends in different topics may enable researchers to gain a deeper understanding of how their field is evolving and potentially identify what factors may be driving these changes.
Each journal contributed a different number of papers to each field, with some journals publishing more papers in some fields and fewer in others. The NEJM contributed 62.3% of all genetics papers included in this study, while the Lancet contributed 4.9%. Since the NEJM included the largest number of studies, it can be expected to contribute a larger percentage to most topics than other journals, however, the difference in the percentages contributed by Lancet and NEJM cannot be explained solely by differences in the number of papers published. The plot shows that different journals tend to publish a different proportion of papers in certain fields when compared to other journals, and therefore when researchers attempt to publish in journals, selecting journals that have published in their desired fields may be worth considering.
Even though topic modeling with conventional methods is modestly discussed in the literature8–11 and has produced encouraging results, it has not yet revolutionized research methods and conduct. As far as the authors are concerned, there are no previous studies using BERTopic to model topics in medical literature, the most common uses for BERTopic have been limited to analyzing different topics across social media and other media platforms.12–14 BERTopic and traditional methods (LDA for example) are two different approaches to topic modeling. While LDA has been the traditional method of topic modeling for many years, BERT is a more recent approach and one of the goals of our study was to assess how effective it is in the context of medical literature. One of BERT’s distinguishing features is its ability to use contextual embeddings and bidirectional processing, both of which provide BERTopic with a more comprehensive understanding of the data and allow it to better capture the underlying topics,13,15,16 whereas traditional methods utilize a statistical approach and only consider the frequency of words within a document to draw conclusions. BERTopic and its word embeddings can be trained on massive amounts of text data, allowing them to capture patterns in the data that traditional methods cannot. The above-mentioned features are particularly useful in medical research, where the context of words is critical, and the meaning of certain words differs from their use in other fields.
Despite the novelty of the methodology, the study has several limitations that need to be considered before using it. The model could not classify around 20% of the inputted abstracts. However, such a proportion of unclassified abstracts will not affect the overall trend. Even though most classification stages have been automated, it is still necessary to manually classify the extracted topics because the model will provide researchers with five descriptive words for each topic, and they must then give each word cluster a topic name. Finally, while we extracted data from the Web of Science database, one of the well-curated literature databases,17 the trends extracted were based on three years of publishing history in the top four medical journals. Future studies might need to consider a larger number of journals over an extended period beyond the COVID-19 publishing pandemic.18
We demonstrate a novel method of using the BERTopic NLP model to analyze topics discussed in large numbers of articles. The study shows that this methodology can identify the most common topics discussed in medical literature while demonstrating the effectiveness of this approach in identifying and extracting relevant information from a large amount of data. This methodology has the potential to analyze trends in medical literature more efficiently and effectively, potentially enabling physicians and researchers to gain a deeper understanding of how their field is evolving. Furthermore, this study’s methodology can be replicated on a larger scale with more papers, more journals, over a longer period, and over different fields. This study also highlights the importance of further research using BERTopic and other NLP models, especially in fields such as the medical field where it is becoming difficult to overcome the challenges of dealing with large volumes of complex data, as NLP has the potential to revolutionize research methods and conduct.
Saif Aldeen AlRyalat: The University of Jordan, 11942 Amman, Jordan. Mobile: +962-798914594. Email: s.alryalat@ju.edu.jo , saifryalat@yahoo.com
All authors declare that they have no competing interests.